INDI - Documentation
Integrated Nanobody Database for Immunoinformatics (INDI) curates sequences and text metadata associated with heavy-chain only antibodies, alternatively known as Nanobodies, VHH, single-domain antibodies or single-chain antibodies. On this page you will find an explanation of the contents of the database
Table of contents - please click on entries to scroll to the section.
Database Contents
In order to provide a single-point-of entry to Nanobody data in the public domain, we collect information from major sources of deposition of biological sequences. This facilitates access to information in those specific molecule types within heterogenuous biological sequences deposited within the sources. Each of the sources and its antibody context is described briefly below.
Patent Nanobodies. We collect nanobodies that are associated with patent documents in the form of protein listings. We identify the antibody sequences and on the basis of sequence features, patent document text and classification assess whether it contains Nanobodies or not. Each Nanobody sequence from patent is associated with the text metadata of the document it originated from such as patent title, abstract etc. to facilitate text-based searches for sequences associated with specific biological entities (e.g. particular targets).
GenBank Nanobodies GenBank is one of the primary structured sources for the deposition of biological sequences. Nanobodies from this data are identified by antibody/nanobody-like sequences features combined with text mining of the metadata of deposition. Each GenBank Nanobody sequence is associated with its source deposition document which among others includes the source organism, deposition description and source publication which facilitates text-based retrieval of Nanobody sequences associated with certain biological entities (e.g. particular targets).
Structures of Nanobodies The Protein Data Bank (PDB) is the primary public source for three dimensional conformation data on biomolecules. Nanobody sequences from the PDB are identified by the virtue of antibody-nanobody sequence features and text mining of metadata fields associated with particular chains and the entire PDB documents.
Next-Generation Sequencing Nanobodies Next Generation Sequencing now allows us to query the great sequence variability covered by antibody sequences. We identify bioprojects that specifically performed NGS of Nanobodies from camels, llamas or alpacas. Querying and analyzing such datasets provides richer information on the sequence variability of Nanobodies than studying limited germline sequences available.
Manually-curated sequences Oftentimes biological sequences are not deposited in standardized repositories such as GenBank and these can instead be directly in scientific publications and their supplementary material. There currently does not exist a reliable automatic method to identify such sequences. Therefore this category encompasses such Nanobody sequences that we add to our database on the basis of manual curation of scientific publications. Nanobody sequences here are linked to the metadata of publications that they originate from to facilitate text-based retrievals.
Table 1. Metadata entries collected for each data source
| Source | Metadata entries |
|---|---|
| Structures |
|
| Patents |
|
| GenBank |
|
| NGS |
|
| Manual |
|
Data Retrieval
In order to facilitate interaction with Nanobody data in INDI we mapped the most common retrieval tasks. We have divided these among Variable Sequence Search, CDR-3 Search and Text Search. The sequence retrieval methods we developed are antibody/nanobody-specific providing improved retrieval ass opposed to basic biological sequence search as with BLAST. Our text-retrieval functionality was designed to provide an easy piont of entry regardless of the heterogenuous sources and their associated metadata.
Variable Sequence Search
The variable sequence of a Nanobody defines its binding specificity by virtue of the three Complementarity Determining Regions. We implemented a nanobody-specific search facility that identifies the closest full variable sequence in INDI to the query sequence.
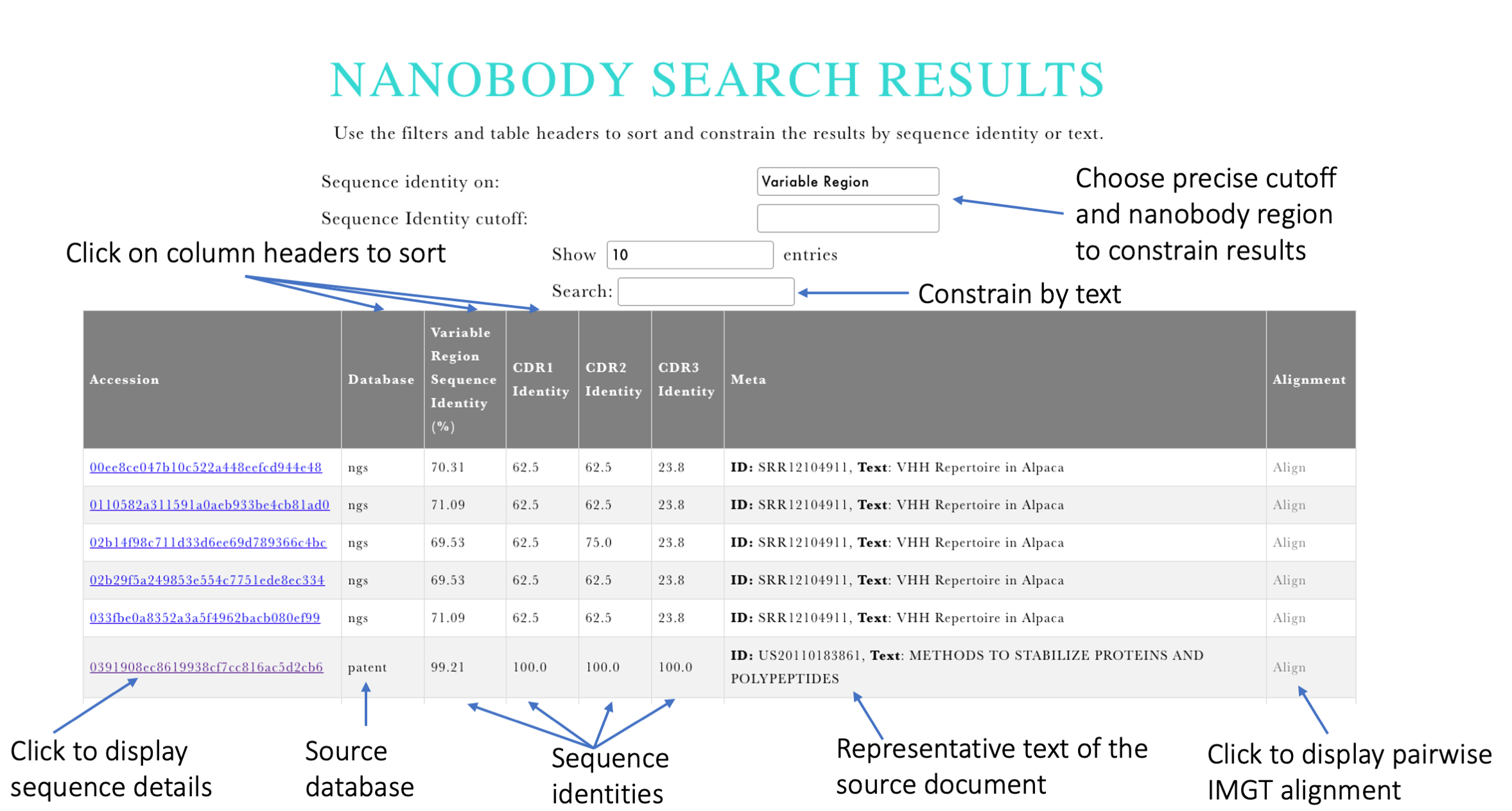
Input: The user is asked to provide a variable region sequence of the nanobody (an example is also provided in the form of therapeutic Nanobody Caplacizumab). Upon clicking Search the system will IMGT-align the query sequence to these in INDI and return the top results sorted by highest sequence identity.
Output: The results give the breakdown of sequence identities of the matches together with the associated text metadata specific to one of our five data sources (Figure 1). Sequence-identity results are broken into the identity on the entire variable region and the individual CDRs to reflect the nanobody-specific nature of the search. The table is interactive so the user can sort the results by sequence identity by either clicking table headers or using the dropdown and specifying a precise value for identity. The returned text fields are database-specific and use can use the supplied text search field to constrain the results.
Figure 1. Output of Nanobody variable region search.
CDR-3 Search
CDR-H3 is the most variable of the CDRs and typically carries the biggest proportion of all atomic contacts mediated with the antigen. Therefore, it is desirable to discover sequences of CDR-H3s that are sequence similar to the query, regardless of the identity of the rest of the sequence (e.g. particular germeline or CDR1/CDR2). Our nanobody CDR-H3 search functionality allows the user to retrieve sequences of nanobodies where CDR-H3 is sequence similar to the query.
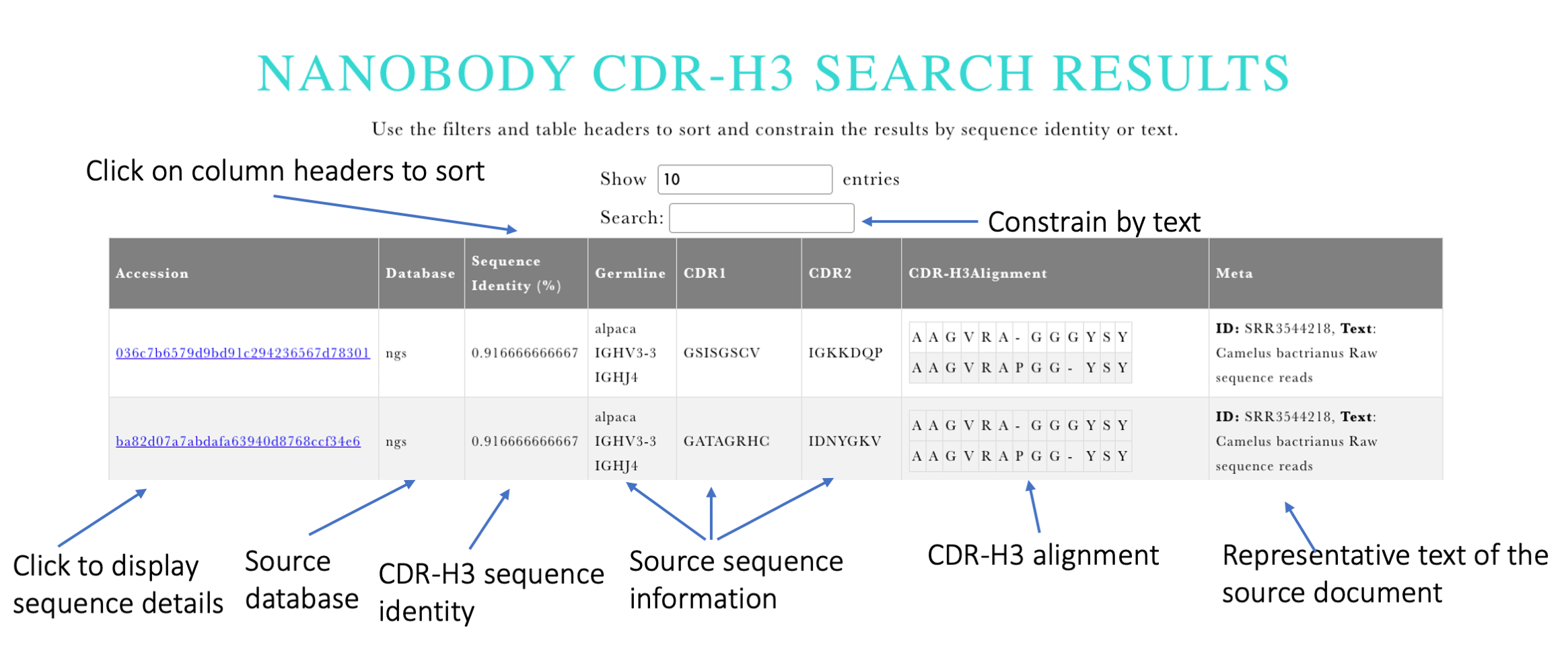
Input: The user is asked to provide a sequence of an IMGT CDR-H3 (an example is also provided). Upon clicking Search the system will align the input CDR-H3 sequence to IMGT CDR-H3 sequences in INDI (not necessarily the same length). The top results given by sequence identity are returned.
Output: The results give the sequence identities of the aligned CDR-H3s and information on the full sequences where they originate from (Figure 2). The table is interactive so the results can be sorted by CDR-H3 identity. The precise alignment between the query and result CDR-H3s are also provided. Each result is accompanied by the meteadata entries associated with the given sequence and specific to the particular database. The results can be constrained based on metadata by entering matching keywords to the text search field above the results table.
Figure 2. Output of Nanobody CDR-H3 search.
Text Search
Nanobody sequences are deposited in diverse repositories based on their sources. They are all associated with rich textual annotations that are source-specific. Text annotation heterogeneity exists not only across source databases but also within them- without stringent deposition rules and guidelines on conveying text information it is up to the authors where they include crucial information. For instance, in the structural entries keyword 'nanobody' can be found either in the title of the entire PDB entry or in the fasta header for a particular chain. Therefore so as to solve the heterogeneity problem with text metadata search we implemented a text index created on all the metadata fields in all the databases. In this fashion, searching for particular keywords (e.g. possible targets) has a single and simple point of entry rather than being burdened with lengthy field-specific forms.
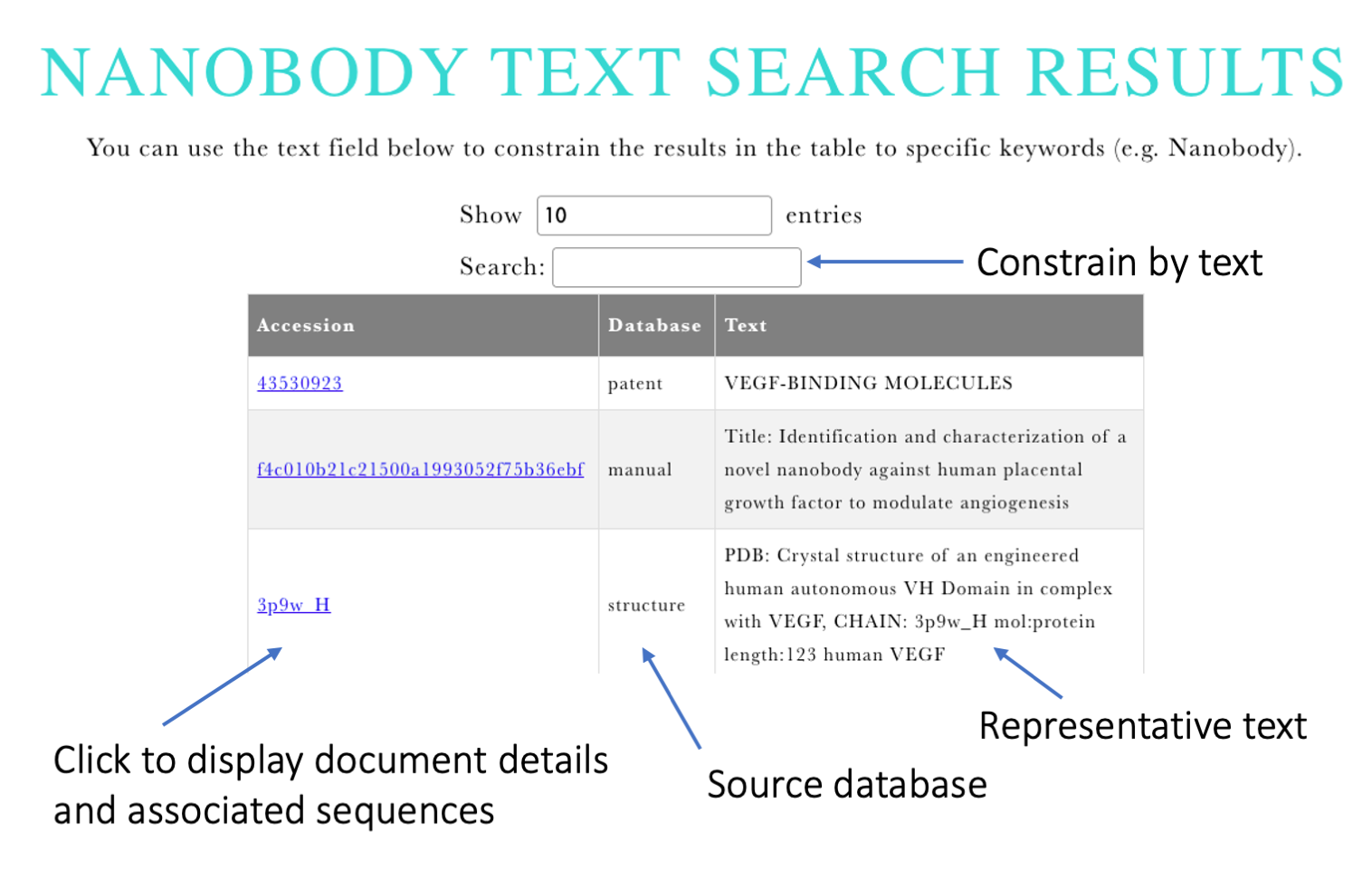
Input: The user is asked to provide keywords, which can be biological entities (e.g. VEGF), commercial organizations (e.g. Ablynx) or diseases (e.g. COVID) - however the search is not constrained to those categories and is performed on all text fields.
Output: The results are presented in an interactive table showing representative text of the entries where the keywords were identified. Search field above the table can be used to further constrain the results. Individual entries can be explored by clicking on the document identifier. This will carry the user to the page where the matching document information is displayed together with its associated nanobody sequences.
Figure 3. Output of Nanobody text search.
Contact
INDI was developed by NaturalAntibody. If you have any questions regarding the contents of this database, found errors, have suggestions for improvements or similar, please get in touch at contact@naturalantibody.com.
Terms of Use
INDI is free for all to use. We ask the users to use the system reasonably as excessive programmatic querying of th data can result with service disruption to other users - for such endeavors we rather suggest to get in touch with us contact@naturalantibody.com
