Patented Antibody Database (PAD) - Documentation
The Patented Antibody Database comprises antibody sequences found in patent documents from primary sources (USPTO, WIPO) and third parties (DDBJ, EBI). Current release of the database is October 2020. The database currently encompasses ca 267,722 antibody chains (148,774 heavy chains and 118,948 light chains) from 19,037 patent families.
The database is aimed at antibody engineers in industry and academia to help navigate the landscape of the protected antibodies. Below we describe the contents of the database, how to interact with it and how to use the search functionality.
Database Contents
PAD curates the antibodies found in the sequence entries reported with patent applications. The database encompasses the antibody chains where we could identify all the variable region fragments (four framework regions and four CDR regions) and containing only the 20 canonical amino acids. For each antibody sequence we record the following values:
Antibody sequence The variable region sequence that we could find in a patent document. This was either from protein submissions or after translation from nucleotideds.
Identified Molecules We performed Named Entity Recognition on the patent documents to identify the molecules that could be associated with the antibodies in a given patent document (e.g. as targets).
Patent Family. The patent documents the sequence was found in.
Patent Metadata. Titles and applicants from the corresponding patent documents.
Web-based patent antibody sequence search
We facilitate the interaction with the data in PAD by offering a sequence-search functionality available here. Below we describe the input and output pages and how to use them.
Input: The user is asked to provide a sequence of an antibody heavy or light chain (an example is given). Upon clicking Search the system will align the input sequence to the patented sequences according to the IMGT scheme. Top-50 results by sequence identity are displayed in the output screen.
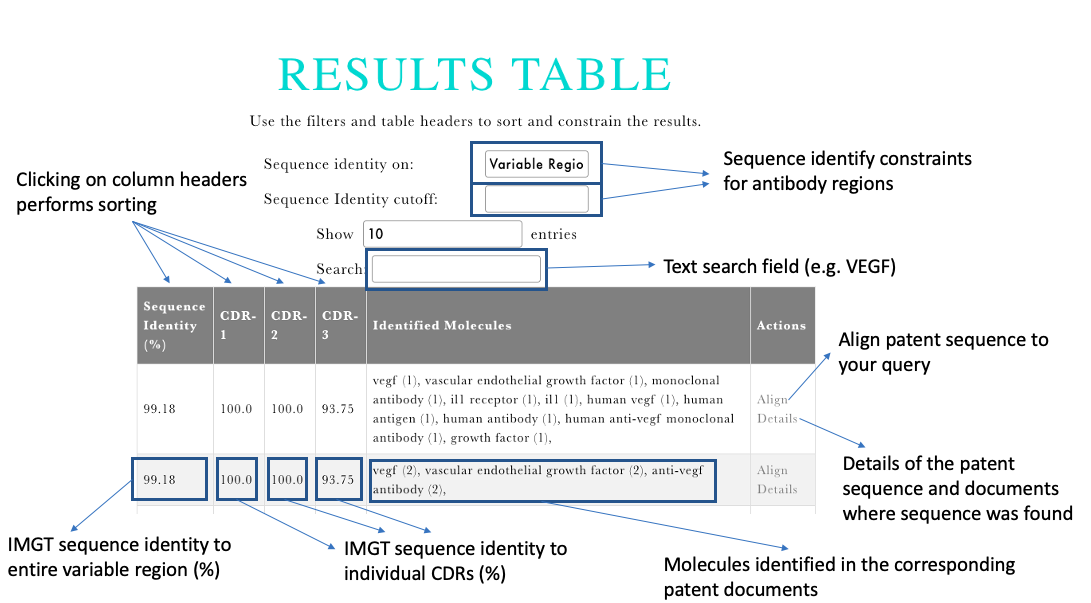
Output: The output displays the top 50 results sorted by the IMGT sequence identity to your query sequence. Descriptions of the resulting fields are shown in the figures below.
Sequence Identity. The main Results Table shows the sequences with their sequence identities for the entire variable region and individual CDRs (IMGT). The sequenes can be sorted by the sequence identities by either clicling on the table headers or choosing a specific region in the drop-down menu above the table and specifying the sequence identity.
Identified Molecules. The Identified Molecules show the results of Named Entity Recognition results from the patent documents associated with the given sequence. The text search field above the Results Table can be used to constrain the results only to the input search phrase (e.g. vegf).
Actions Clicking on the Alignment button, will show the query aligned to the input sequence, with the framework regions and CDRs highlighted. Clicking on the Details button will display the information on the particular sequence from patents and information on the documents where it was identified.
License
Patented Antibody Database and all its contents were developed by NaturalAntibody and are released under CC-BY-NC license, so in lay terms reuse is permitted so long its non-commercial. If you would like to use these data for commercial purposes or install it in-house, please contact konrad@naturalantibody.com for details.
